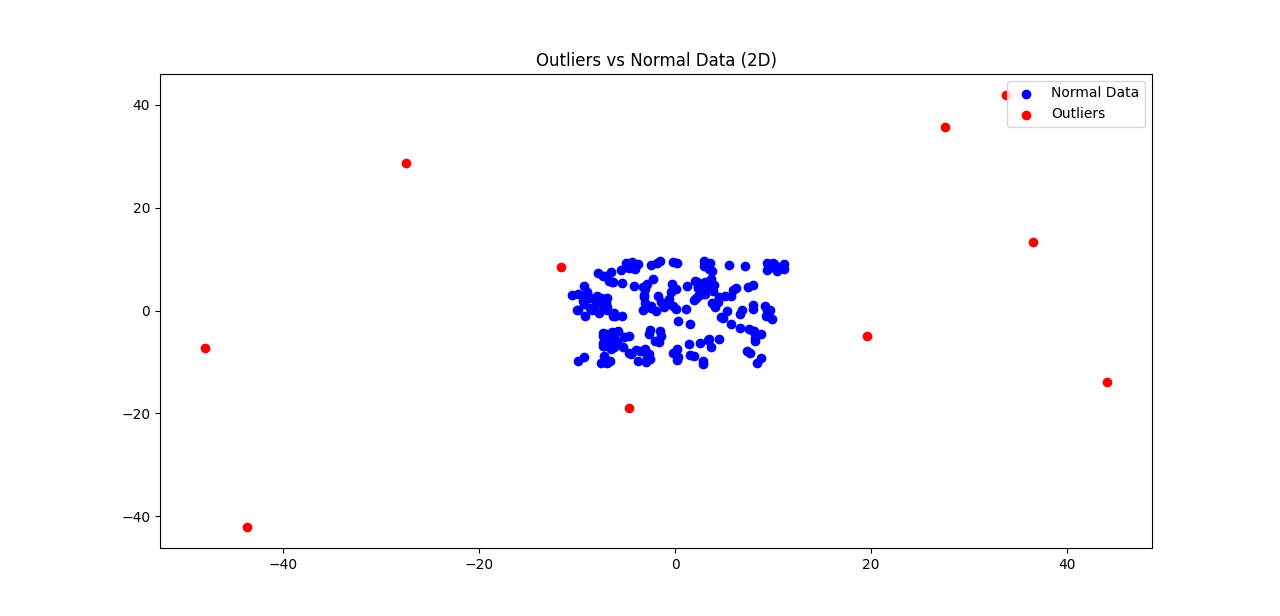
Usage Example for Random 2D Data
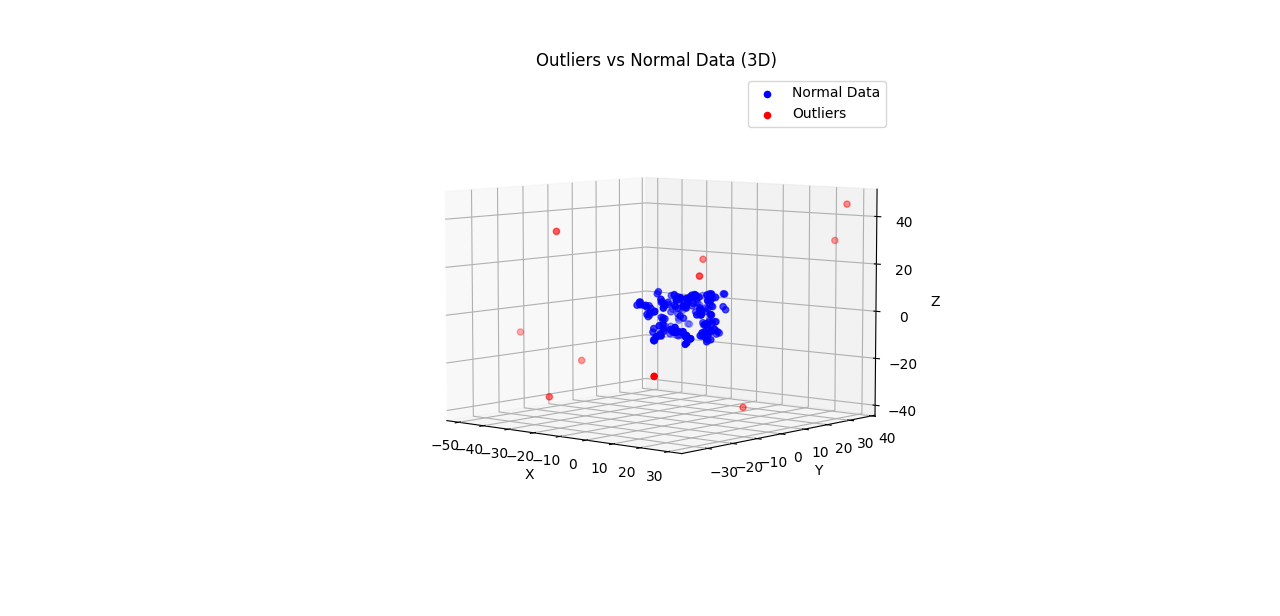
Usage Example for Random 3D Data
Python library for Multi-Dimensional Data Outlier/Anomaly Detection Machine Learning algorithm.
pip install mdodgit clone https://github.com/mddod/mdod.gitcd mdodpython setup.py installPS D:\mdod> python testmdodmodelv3.pyData generation failed after 1000 attempts. Scaled data to fit within 1.0.Generate data: 1000 Sample, 2 Dimension, 150 OutliersGenerated data has been saved to 'generated_data.csv'Use sampling rate: 0.05 MDOD Decision Score (Top 10):[-18.31559252 -19.31589488 -18.95489266 -19.27465188 -19.38318373 -19.02271112 -19.5686593 -19.2839105 -18.60204328 -18.33769728]MDOD Predicted Labels (Number of Anomalies): 150MDOD AUC: 1.0MDOD Runtime: 0.002752 secondsMDOD Confusion Matrix:[[850 0] [ 0 150]]MDOD Precision: 1.0000MDOD Recall: 1.0000MDOD F1-Score: 1.0000LOF Decision Score (Top 10):[1.25375406 1.01080138 1.11113192 1.0219336 1.03138014 1.10677047 0.96855755 0.99321085 1.17408394 1.30132643]LOF Predicted Labels (Number of Anomalies): 150LOF AUC: 1.0LOF Running time: 0.011232 secondsLOF Confusion Matrix:[[850 0] [ 0 150]]LOF Precision: 1.0000LOF Recall: 1.0000LOF F1-Score: 1.0000Comparison results:Spearman correlation coefficient: 0.9244 (p-value: 0.0000)MDOD AUC: 1.0000, LOF AUC: 1.0000Algorithm Complexity Comparison (Empirical Running Time):MDOD 0.002752s vs LOF 0.011232sTest Data Comparison (Top 10 Rows): Feature_1 Feature_2 True_Label MDOD_Score MDOD_Label LOF_Score LOF_Label0 0.236384 0.463173 0.0 -18.315593 0 1.253754 01 -0.231236 0.046397 0.0 -19.315895 0 1.010801 02 0.089341 0.378934 0.0 -18.954893 0 1.111132 03 -0.289616 0.078549 0.0 -19.274652 0 1.021934 04 0.252438 -0.010620 0.0 -19.383184 0 1.031380 05 0.374958 0.164868 0.0 -19.022711 0 1.106770 06 0.037580 0.167901 0.0 -19.568659 0 0.968558 07 -0.230299 -0.145481 0.0 -19.283910 0 0.993211 08 0.003694 0.429171 0.0 -18.602043 0 1.174084 09 -0.449619 -0.343795 0.0 -18.337697 0 1.301326 0The complete data has been saved to 'test_data_comparison.csv'Test output data has been saved to 'test_output_data.csv'libpng warning: iCCP: cHRM chunk does not match sRGBlibpng warning: iCCP: cHRM chunk does not match sRGBDownload testmdod_simple_example_en.py read data from csv file.
D:\mdodtest>D:\mdodtest>D:\mdodtest>D:\mdodtest>py testmdod_simple_example_en.pyNumber of features: 2Number of samples: 1000Number of outliers: 150Data saved to: testmdod_simple_example_output_data.csvSampling rate used: 0.05MDOD runtime: 0.0797 secondsMDOD decision scores (top 10 - highest values):[-6.62289984 -6.77322132 -6.92891522 -7.18871113 -7.23625849 -7.24312221-7.31773667 -7.32676678 -7.33133378 -7.33982723]MDOD decision scores (bottom 10 - lowest values):[-19.84866083 -19.82038598 -19.82010554 -19.80062169 -19.80051546-19.76201034 -19.76181435 -19.75729718 -19.75229983 -19.74520579]parameters of MDOD include: norm_distence: The distance of the virtual dimension (the default is 1.0, which affects the similarity calculation).top_n: Consider the number of nearest points used for statistics (select the appropriate number for outlier identification and distinction).Contamination: The set abnormal ratio (the default is 0.1, which is used for threshold calculation).sampling_rate: sampling rate (0~1, default 1.0, low value accelerates calculation).random_state: random seed.

Visit GitHub(github.com/mddod/mdod) or PyPI(pypi.org/project/mdod/) for more information.